Our Research
Gut microbiomes through the lens of cultivation
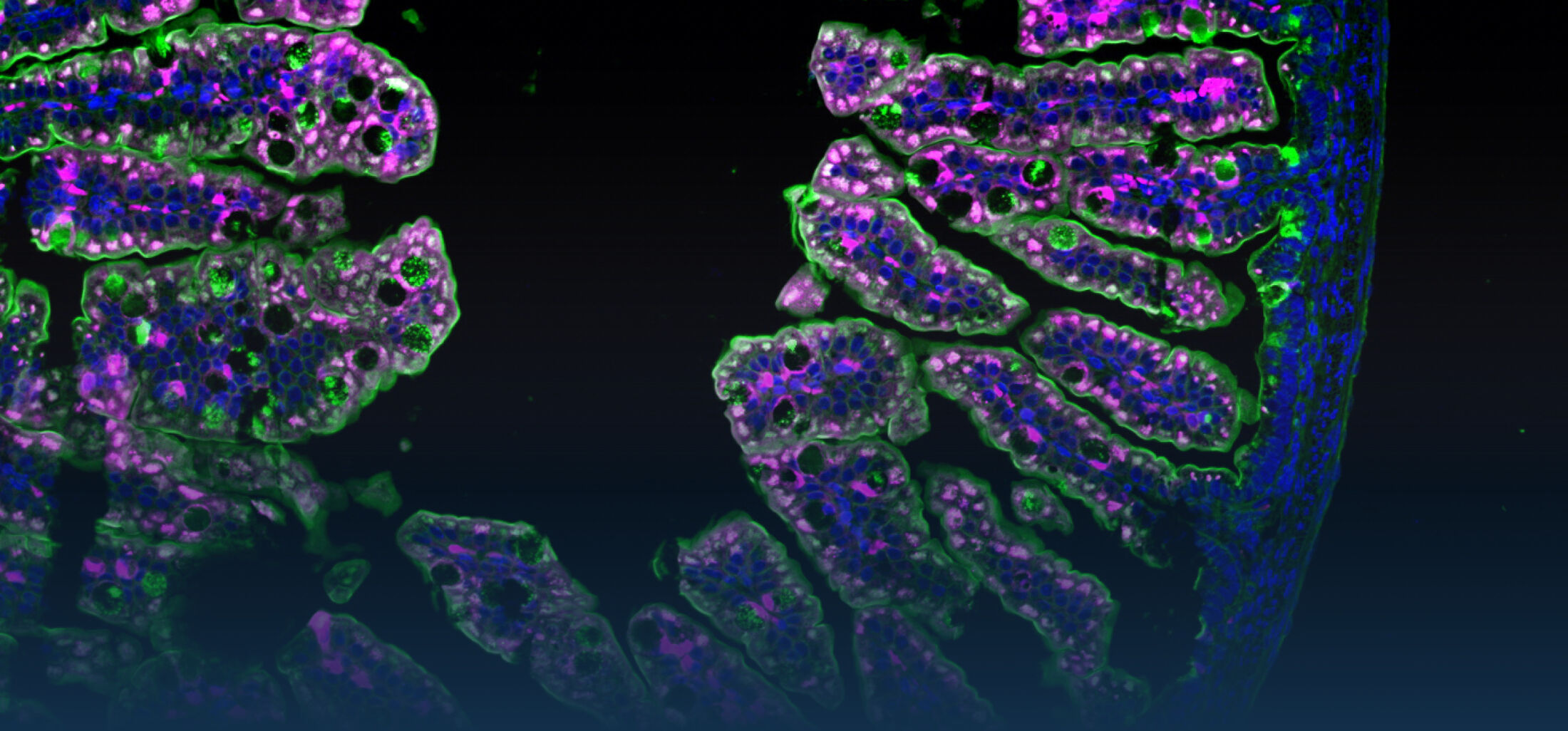
The diversity of bacteria on earth is tremendous. In the intestine of mammals, one to two thirds of prokaryotic diversity remain unexplored (i.e., strains of many taxa have never been isolated and/or described). This pool of unknown diversity represents a substantial phylogenetic hole and most of all an opportunity to discover novel bacterial functions. An important part of our work is dedicated to the cultivation and description of new taxa from the intestine of humans and animals. Because intestinal microbiomes have co-evolved with their host species, we put effort in establishing comprehensive collections of publicly available bacterial strains in a host-specific manner (in collaboration with the Leibniz-Institute DSMZ). These collections serve as a foundation for experimental work to study the ecology of gut microbes as well as microbe-host interactions using in vitro (continuous culture) and in vivo (gnotobiology) systems. We also currently work on an automated workflow for anaerobic cultivation (iSOMiC platform) to generate personalized collections of gut microbes.
Microbe-host interactions
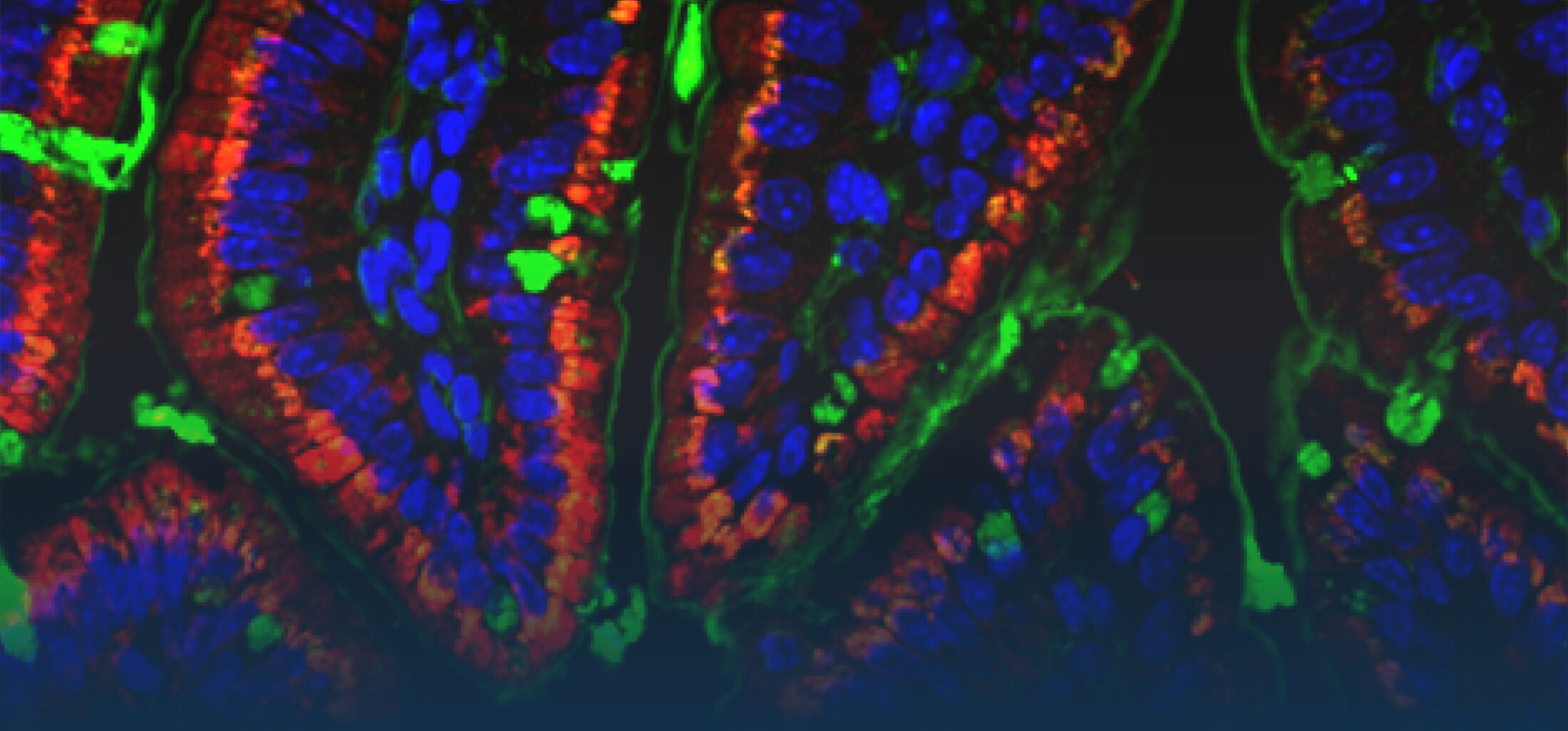
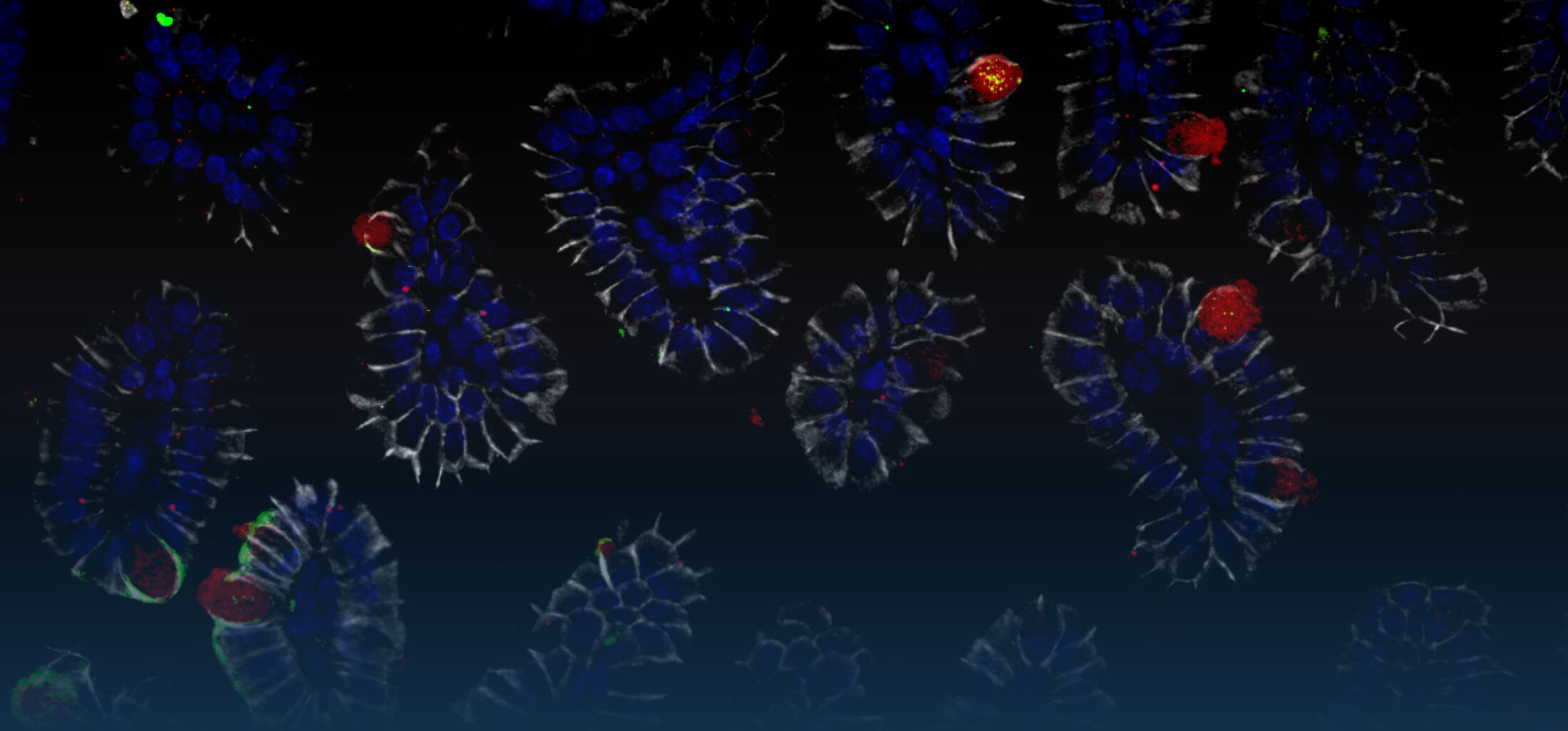
Because intestinal bacteria constantly interact with dietary factors and host cells and because they produce a myriad of bioactive molecules, they are known to influence the physiology of their host and can be implicated in the development of chronic diseases. The last decade of microbiome research has generated many descriptive data demonstrating shifts in gut microbiome structure and functions associated with diseases. Effort is now required to strengthen our knowledge about molecular mechanisms underlying microbe-host interactions. We are particularly interested in studying the effects of gut microbiota members on the metabolism of lipids, with primary focus on bile acids. Model species within the Coriobacteriales but also other secondary bile acid- and lipase-producing taxa and their impact on liver physiology, metabolic disturbances, and the development of colorectal cancer are under investigation. In addition, we study the biochemistry of small SCIFF proteins produced by own isolates of Gram-positive bacteria and their role within the ecosystem.


Intestinal microbes, diet & health
As gut bacteria are known to metabolize compounds from the diet and thereby modulate their bioavailabiltiy and bioactivities, we have investigated the conversion of dietary polyphenols and identified several bacteria responsible for the production of bioactive metabolites. Our current research focuses on the conversion of dietary and host-derived lipids.
Diet can modulate the gut microbial ecosystem and the use of molecular techniques has been really helpful in dissecting diet-microbiota interactions in a culture-independent manner and assessing their importance for health regulation. We have been using high-throughput sequencing to identify microbiome signatures associated with specific dietary components and host pathologies. The culture resources aforementioned allow us to test the generated hypotheses in experimental models.
Selected publications
- Afrizal A, Jennings SAV, Hitch TCA, Riedel T, Basic M, Panyot A, Treichel N, Hager FT, Wong EO, Wolter B, Viehof A, von Strempel A, Eberl C, Buhl EM, Abt B, Bleich A, Tolba R, Blank LM, Navarre WW, Kiessling F, Horz HP, Torow N, Cerovic V, Stecher B, Strowig T, Overmann J, Clavel T. Enhanced cultured diversity of the mouse gut microbiota enables custom-made synthetic communities. Cell Host Microbe. 2022 Nov 9;30(11):1630-1645.e25. https://doi.org/10.1016/j.chom.2022.09.011
- Afrizal A, Hitch TCA, Viehof A, Treichel N, Riedel T, Abt B, Buhl EM, Kohlheyer D, Overmann J, Clavel T. Anaerobic single-cell dispensing facilitates the cultivation of human gut bacteria. Environ Microbiol. 2022 Sep;24(9):3861-3881. https://doi.org/10.1111/1462-2920.15935
- Hitch TCA, Riedel T, Oren A, Overmann J, Lawley TD, Clavel T. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun. 2021 May 20;1(1):16 https://doi.org/10.1038/s43705-021-00017-z
- Wylensek D, Hitch TCA, Riedel T, Afrizal A, Kumar N, Wortmann E, Liu T, Devendran S, Lesker TR, Hernández SB, Heine V, Buhl EM, M D'Agostino P, Cumbo F, Fischöder T, Wyschkon M, Looft T, Parreira VR, Abt B, Doden HL, Ly L, Alves JMP, Reichlin M, Flisikowski K, Suarez LN, Neumann AP, Suen G, de Wouters T, Rohn S, Lagkouvardos I, Allen-Vercoe E, Spröer C, Bunk B, Taverne-Thiele AJ, Giesbers M, Wells JM, Neuhaus K, Schnieke A, Cava F, Segata N, Elling L, Strowig T, Ridlon JM, Gulder TAM, Overmann J, Clavel T. A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat Commun. 2020 Dec 15;11(1):6389. https://doi.org/10.1038/s41467-020-19929-w
- Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martínez I, Just S, Ziegler C, Brugiroux S, Garzetti D, Wenning M, Bui TP, Wang J, Hugenholtz F, Plugge CM, Peterson DA, Hornef MW, Baines JF, Smidt H, Walter J, Kristiansen K, Nielsen HB, Haller D, Overmann J, Stecher B, Clavel T. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016 Aug 8;1(10):16131. https://doi.org/10.1038/nmicrobiol.2016.131
Further reading
- Jennings SAV, Clavel T. Synthetic Communities of Gut Microbes for Basic Research and Translational Approaches in Animal Health and Nutrition. Annu Rev Anim Biosci. 2023 Nov 14; https://doi.org/10.1146/annurev-animal-021022-025552
- Pabst O, Hornef MW, Schaap FG, Cerovic V, Clavel T, Bruns T. Gut-liver axis: barriers and functional circuits. Nat Rev Gastroenterol Hepatol. 2023 Jul;20(7):447-461; https://doi.org/10.1038/s41575-023-00771-6
- Hitch, Hall, Walsh, Leventhal, Slack, de Wouters, Walter, Clavel (2022) Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol, https://doi.org/10.1038/s41385-022-00564-1
- Clavel, Horz, Segata, Vehreschild (2021) Next steps after 15 years of human gut microbiome research. Microbial Biotechnology, http://doi.org/10.1111/1751-7915.13970
- Daniel, Hauner, Hornef, Clavel (2021) Allulose in human nutrition: the knowns and the unknowns. Br J Nutr, https://doi.org/10.1017/S0007114521003172
- Hitch, Afrizal, Riedel, Kioukis, Haller, Lagkouvardos, Overmann, Clavel (2021) Recent advances in culture-based gut microbiome research. Int J Med Microbiol 311:151485 ; https://doi.org/10.1016/j.ijmm.2021.151485
- Clavel (2019) Microbiote intestinal, lipides alimentaires et maladies métaboliques (in French). Cahiers de Nutrition et Diététique 50:237; https://doi.org/10.1016/j.cnd.2019.07.005
- Clavel, Ecker (2018) Microbiome and diseases – Metabolic disorders, In The Gut Microbiome in Health and Disease, Haller D (Ed.), Springer, ISBN 978-3-319-90544-0; https://doi.org/10.1007/978-3-319-90545-7_16
- Clavel, Lagkouvardos, Stecher (2017) From complex gut microbial communities to model bacterial consortia via cultivation. Curr Opin Microbiol 38:148; https://doi.org/10.1016/j.mib.2017.05.013
- Clavel, Gomes-Neto, Lagkouvardos, Ramer-Tait (2017) Deciphering interactions between the gut microbiota and the immune system via microbial cultivation and minimal microbiomes. Immunol Reviews 279:8; https://doi.org/10.1111/imr.12578
- Clavel, Hörmannsperger (2017) Darmmikrobiom des Menschen: Status quo und Perspektiven (in German), In Pädiatrie up2date 12 (04), 335-349; https://doi.org/10.1055/s-0043-115278
- Clavel, Lagkouvardos, Hiergeist (2016) Microbiome sequencing: challenges and opportunities for molecular medicine. Expert Rev Mol Diagn 16:795; https://doi.org/10.1080/14737159.2016.1184574
- Clavel, Lagkouvardos, Blaut, Stecher (2016) The mouse gut microbiome revisited: from complex diversity to model ecosystems. Int J Med Microbiol 306:316; https://doi.org/10.1016/j.ijmm.2016.03.002
- Clavel, Mapesa (2013) Phenolics in human nutrition: importance of the intestinal microbiome for isoflavone and lignan bioavailability, In Handbook of Natural Products; Ramawat, Merillon (Eds); Elsevier, ISBN 978-3-642-221-43-9/44-6; https://doi.org/10.1007/978-3-642-22144-6_94
- Hörmannsperger,* Clavel,* Haller (2012) Gut matters: intestinal microbial ecology in allergic diseases. J Allergy Clin Immunol 129:1452; https://doi.org/10.1016/j.jaci.2011.12.993
- Clavel, Doré, Blaut (2007) Bioavailability of lignans in human subjects. Nutr Res Rev 19:187; https://doi.org/10.1017/S0954422407249704
- Blaut, Clavel (2007) Metabolic diversity of the intestinal microbiota: Implications for health and disease. J Nutr 137:751S; https://doi.org/10.1093/jn/137.3.751S