Our Research
Our research focuses on the biophysics of host-microbe interactions, using and extending quantitative methods to assess how microbes navigate the host environment.
Motivations
An estimated half of all bacteria can swim by rotating one or more flagella. Most are also capable of biasing this swimming to ascend or descend a chemical gradient in a process called chemotaxis. There are very strong incentives to think that motility and chemotaxis are important in the early steps of inflammation and/or infection, allowing bacteria to breach the mucus barrier that protects our mucous membranes.
Navigation capabilities have been extensively studied in the peritrichously flagellated model organism E. coli in buffer, but much less attention has been paid to:
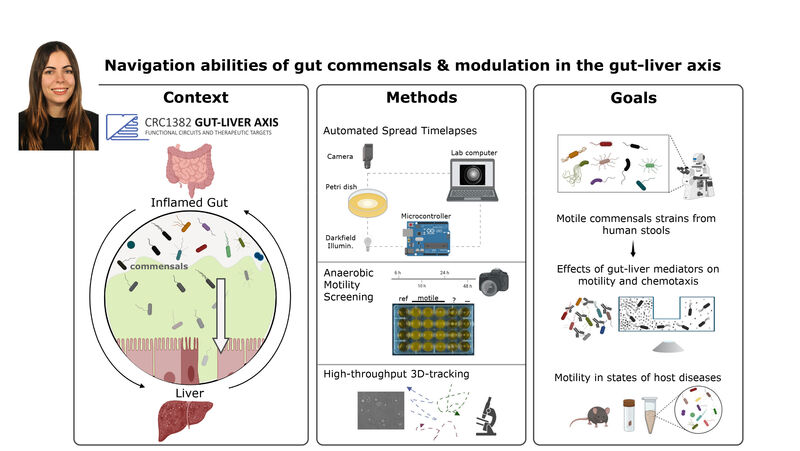
(1) the diversity of motility in other organisms: We ask, for example, how the wide variety of bacterial commensals swim in our gut, and how their motility and chemotaxis may be involved in intestinal inflammatory processes.
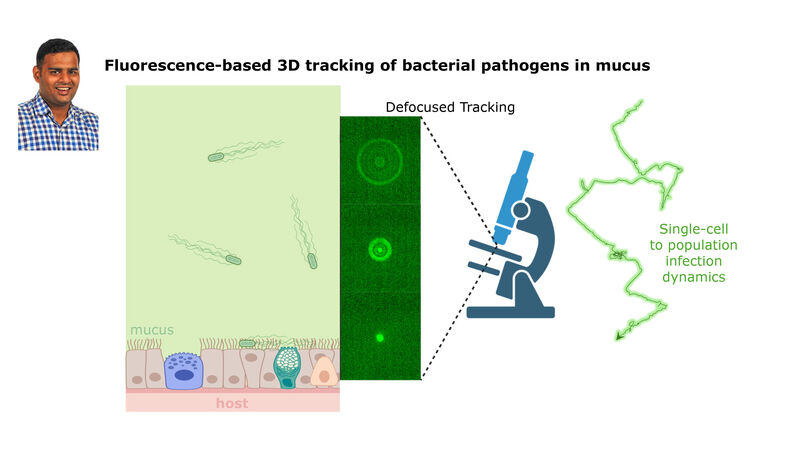
(2) the diversity of motility in viscous or porous environments that more closely mimic the complexity of the host environment: Our current 3D tracking method (see methodological pillars) allows us to track bacteria in buffer, but also in viscous media or agar hydrogels. We are currently developing another bacterial 3D tracking method to assess how pathogens swim in real mucus, from the lumen to the epithelial cell.
Overall we aim to explore the motility and chemotaxis of bacterial pathogens and commensals as a novel and profoundly interdisciplinary angle in the field of host-microbe interactions.
The three original methodological pillars of our work
Figure – Quantitative methods to address motility, chemotaxis and growth, altogether or independently. (A) A standard soft agar plate screening assay: petri dishes filled with soft (0.15-0.3%) agar are inoculated with bacteria. Over time, the motile bacteria spread outwards in the soft agar, motivated by nutrient gradients they themselves create by locally consuming them, creating visible white ring waves. (B) Direct characterization of motility in bulk by the mean of a high-throughput 3D-tracking technique, based on phase-contrast imaging (Taute et al., Nat. Commun., 2015). We can obtain thousands of 3D trajectories of swimming bacteria in minutes, without any staining. The trajectories obtained are analyzed in a semi-automated fashion to determine their main characteristics (speed, type of motility, turning frequency, diffusivity…), and to compare different strains and/or mutants. (C) A multiscale chemotaxis assay we developed (Grognot et al., Commun. Biol., 2021). It makes use of method B to track bacteria, within a commercially available microfluidic device where a linear gradient of our choice is created. This method gives a direct measurement of chemotaxis, the chemotactic drift (average speed at which a population climbs or descends a gradient), while addressing how individuals bias their motility to achieve it.